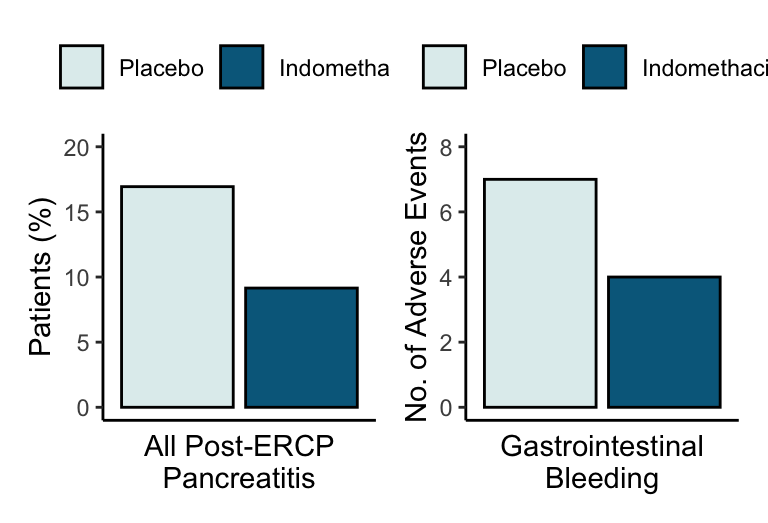
Incidence of the Primary and Secondary End Points and Adverse Events
Bar plots.
── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.4
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.0 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.1
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
library(medicaldata)
library(patchwork)
# A - moderate to severe missing
plot_a <- indo_rct |>
count(rx, outcome) |>
group_by(rx) |>
mutate(p = n / sum(n)) |>
filter(outcome == "1_yes") |>
ungroup() |>
mutate(
rx = case_match(
rx,
"0_placebo" ~ "Placebo",
"1_indomethacin" ~ "Indomethacin"
),
rx = fct_relevel(rx, "Placebo", "Indomethacin"),
p = p*100
) |>
ggplot(aes(x = rx, y = p, fill = rx)) +
geom_col(color = "black") +
scale_fill_manual(
values = c(
"Placebo" = "azure2",
"Indomethacin" = "deepskyblue4"
)
) +
theme_classic() +
theme(
legend.position = "top",
axis.text.x = element_blank(),
axis.ticks.x = element_blank()
) +
ylim(0, 20) +
labs(
x = "All Post-ERCP\nPancreatitis",
y = "Patients (%)",
fill = NULL
)
# renal failure missing
plot_b <- indo_rct |>
filter(bleed == 1) |>
mutate(
rx = case_match(
rx,
"0_placebo" ~ "Placebo",
"1_indomethacin" ~ "Indomethacin"
),
rx = fct_relevel(rx, "Placebo", "Indomethacin")
) |>
count(rx, bleed) |>
ggplot(aes(x = rx, y = n, fill = rx)) +
geom_col(color = "black") +
scale_fill_manual(
values = c(
"Placebo" = "azure2",
"Indomethacin" = "deepskyblue4"
)
) +
theme_classic() +
theme(
legend.position = "top",
axis.text.x = element_blank(),
axis.ticks.x = element_blank()
) +
ylim(0, 8) +
labs(
x = "Gastrointestinal\nBleeding",
y = "No. of Adverse Events",
fill = NULL
)
plot_a + plot_b
Incidence of the Primary and Secondary End Points and Adverse Events